

people | alumini | review articles | google scholar | software | plain english
 Chromosome Segregation
Chromosome Segregation
Our lab works to understand how human cells, eggs and embryos ensure accurate chromosome segregation.
This is important becuase errors can lead to aneuploidy (having the wrong number of chromosomes)
which is implicated in cancer development, developmental syndromes (i.e. Downs) and recurrent pregnancy loss.
A key focus for us is the kinetochore, one of the most complicated pieces of machinery in the eukaryotic cell. Formed from >100 protein components which self-assemble into a multi-megadalton-sized attachment site for the tips of 20 dynamic microtubules. Kinetochores must capture microtubules, support and monitor proper attachment and, ultimately, generate the forces nessessary for chromosome segregation.
Approaches in the McAinsh lab include live-cell microscopy, computational image analysis, mathematical modelling and in vitro reconstitution.
 new: Kinetochoe architecture
new: Kinetochoe architecture
We reveal how the human kinetochores are able to distinguish between changes in microtubule attachment and tension.
>> Roscioli E, Germanova TE, Smith CA, Embacher PA, Erent M, Thompson AI, Burroughs NJ, McAinsh AD. Cell Reports., 2020
| [Open Access]
 new: Corona-control
new: Corona-control
Phil reveals how the kinetochore protein CENP-F helps hold onto microtubules with one hand, while limiting the motorised stripping of corona compenents with the other
>> Auckland P, Roscioli E, Coker H, McAinsh AD. J. Cell Biol., 2020
| [Open Access]
 Kinetochore reconstitution
Kinetochore reconstitution
How Ndc80 and CENPO complexes work together in the kinetochores; Experiments by Phil and Muriel and nice collaboration with Musacchio and Raunser labs
>> Pesenti ME, Prumbaum D, Auckland P, Smith CM, Faesen AC, Petrovic A, Erent M, Maffini S, Pentakota S, Weir JR, Lin Y-C,Raunser S,
McAinsh AD and Musacchio. Molecular Cell, 2018
| [Open Access]
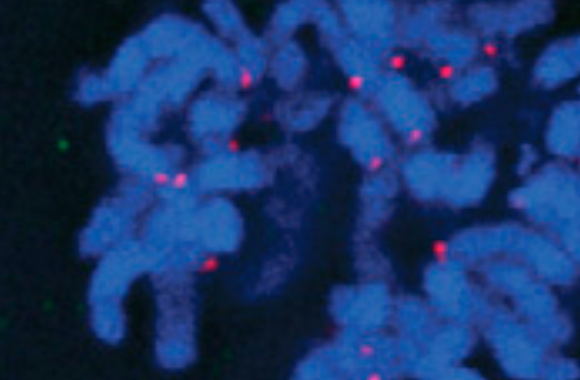 checkpoint suprise
checkpoint suprise
Here we show using genome editing how BUB1 is not essential for the spindle checkpoint in human cells; collaboration with Millar lab
>> Currie CE, Mora-SantosM, Smith CA, McAinsh AD and Millar JBA. Current Biology, 2018
| [Open Access]
 Centrosome clustering mechanisms
Centrosome clustering mechanisms
Collaboration with Susana Godinho - includes tracking of centrosome dynamics by Chris
>> Rhys AD, Monteiro P, Smith C, Vaghela M, Arnandis T, Kato T, Leitinger B, Sahai E, McAinsh A, Charras G, Godinho SA. J. Cell Biol, 2017
| [Open Access]
 More on the Ska complex - Phosphoregulation by Mps1
More on the Ska complex - Phosphoregulation by Mps1
Collaboration with Prasad Jallepalli - includes Hauke's cool in vitro reconstitution of Ska complex on dynamic microtubules.
>> Maciejowski J, Drechsler H, Grundner-Culemann K, Ballister ER, ... McAinsh AD, Jallepalli PV. Dev. Cell. 2017 Dev Cell. 2017 41:143-156
| [Open Access]
 Load-bearing by the Ska complex
Load-bearing by the Ska complex
Phil Auckland finds that kinetochres mature and this is important to prevent premature detachment fromt he mitoic spindle.
>> Auckland, P., Clarke I., Royle S.J. and McAinsh A.D. J. Cell Biol. 2017 J Cell Biol. 2017 Jun 5;216(6):1623-1639
| [Open Access]
 swivel-eyed kinetochores
swivel-eyed kinetochores
Chris Smith shows how kinetochores are a swivel joint that aids connection to the mitotic spindle.
>> Smith A.C., McAinsh A.D. and Burroughs N.J. eLife 2016 5:e16159
| [Open Access]
 motor slide
motor slide
Hauke Drechsler reveals that Kif15 is a multi-function motor that can rearrange microtubule networks, track plus-ends and control dynamics.
>> Drechsler H, McAinsh AD.
Proc Natl Acad Sci U S A. 2016 113:E1635-44
| [Open Access]

Code and User Guides for our software tools and packages can all be found on our CMCB Github page. These include:
KiT (Tracking kinetochores in microscopy movies in 2D or 3D, and in multiple channels)
SiD (Spot Intensity Detector) - unpublished and please use!
