

Research Interests
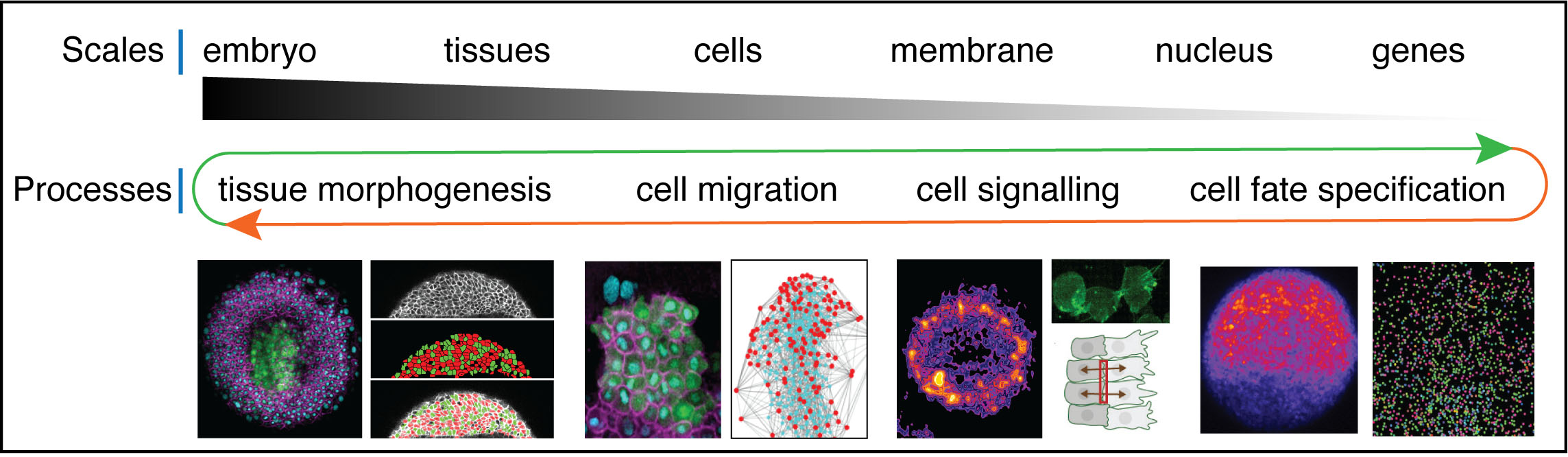
Our research spans over multiple scales of organization: we explore biochemical and mechanical signalling pathways at tissue, cellular and gene expression levels with the ultimate aim to link these events in space and time. We focus our efforts on developmental processes that regulate tissue and organ formation during early zebrafish development including tissue morphogenesis, cell fate specification and tissue patterning, collective cell migration, cell adhesion, nuclear mechanics and gene expression.
Research Methods
Our research encompasses methodologies and technologies such as:
- quantitative live imaging (confocal, multiphoton, light sheet microscopy)
- optogenetics
- embryological methods (ISH, transplantations)
- in vitro reconstitution assays (micropatterning, supported lipid bilayers)
- biophysical tools (microfluidics, laser ablation, magnetic tweezers)
- transcriptomics (scRNA sequencing, multiplexed smFISH)
- gene editing (Crispr/Cas9)
- theoretical modelling (collective motion, biomechanics)
Current Collaborators
Our projects are highly interdisciplinary and we are working closely with national and international collaborators including:
Till Brettschneider (Warwick Computer Science) | Computational Image Analysis
Melikhan Tanyeri (Duquesne University, US) | Microfluidics, Microfabrication
Kok Hao Chen (Singapore Genomic Institute) | Transcriptomics
Justin Molloy (Crick); Darius Koester (Warwick Medical School) | Mechanics, Biophysics
Pierre Haas (MPI Dresden); Matthew Turner (Warwick Physics) | Modelling
Karuna Sampath (Warwick Medial School) | Nodal signaling
Anne Straube (Warwick Medical School) | Microtubule dynamics
Funding





